Introducing the qPCR Primer Analyzer ILVO
🧬 Precision Meets Simplicity
In the ever-evolving field of bioinformatics, precision and usability are paramount. That’s why I developed the qPCR Primer Analyzer ILVO, a streamlined yet powerful web-based tool designed to support researchers in evaluating oligonucleotide sequences for qPCR assays.
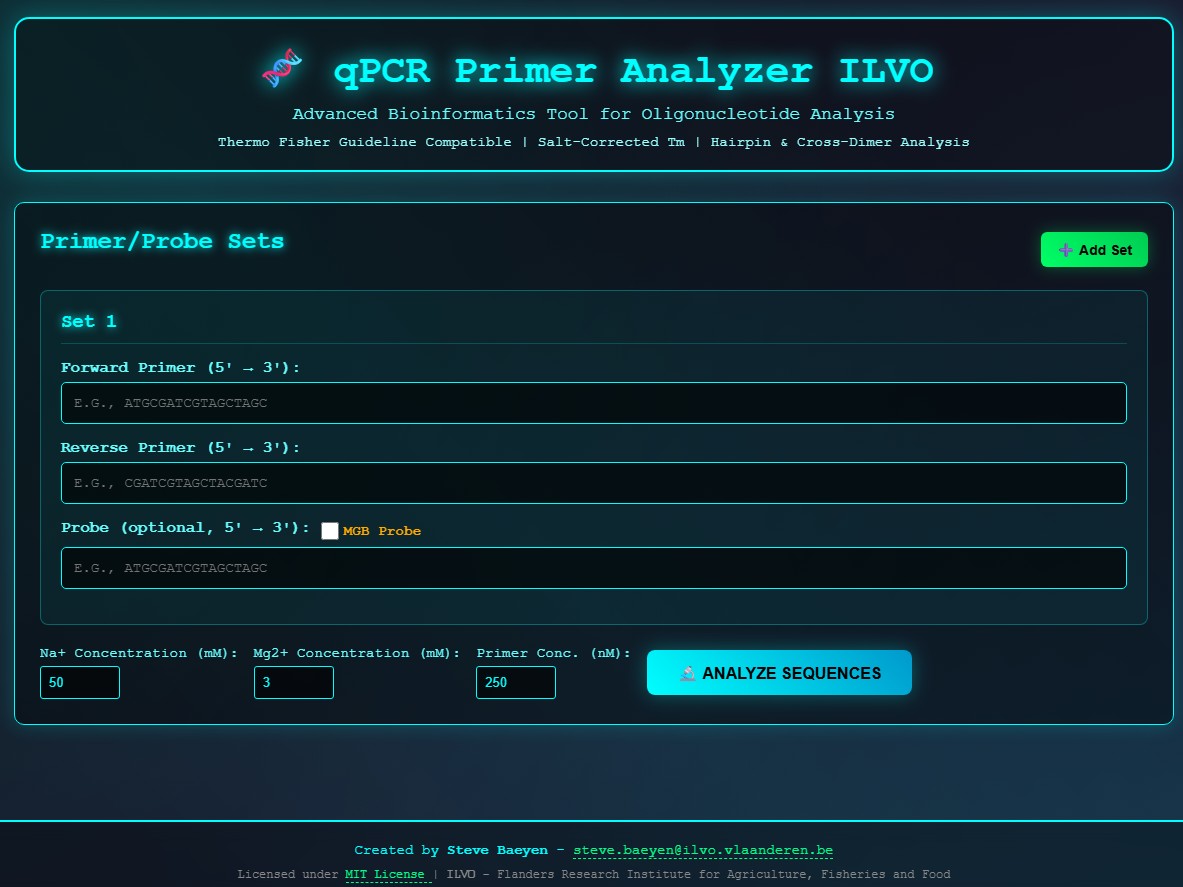
🧪 What It Does
The qPCR Primer Analyzer ILVO helps users assess the quality and compatibility of primer/probe sets with a focus on:
- Salt-Corrected Melting Temperature (Tm): Ensures accurate thermodynamic calculations by factoring in sodium and magnesium concentrations.
- Hairpin and Cross-Dimer Detection: Identifies potential secondary structures that could compromise assay performance.
- Thermo Fisher Guideline Compatibility: Aligns with industry standards for robust and reproducible results.
⚙️ How It Works
Users can input:
- Forward and reverse primers (5’ → 3’)
- Optional probe sequences
- Ion concentrations (Na⁺ and Mg²⁺)
- Primer concentration (nM)
With a single click on 🔬 Analyze Sequences, the tool delivers a comprehensive analysis of the primer set’s thermodynamic properties and structural integrity.
🧑🔬 Built for Researchers, by a Researcher
Created at ILVO – Flanders Research Institute for Agriculture, Fisheries and Food, this tool is open-source and licensed under the https://opensource.org/licenses/MIT. It’s designed to be intuitive for molecular biologists while offering the depth needed by bioinformaticians.
📫 Get in Touch
Have feedback or want to collaborate? Reach out at [steve.baeyen@ilvo.vlaanderen.be]
💻 Clone or download the html file from my repository on ILVO Gitlab
Enjoy Reading This Article?
Here are some more articles you might like to read next: