From Bash to Copilot: My Bioinformatics Terminal Workflow
🧬 Introduction
Welcome to my first blog post on bioinformatics workflows! As a researcher, I often find myself or PhD students juggling with large datasets, remote compute resources, and various command-line tools. My terminal setup has evolved to meet these challenges head-on, allowing me to work efficiently and reproducibly. Efficient command-line workflows are critical in bioinformatics, where data is large, compute is remote, and tools are often UNIX-native. Over time, I’ve developed a terminal setup that allows me to:
- Work on a Windows machine using Linux tools via WSL
- Manage remote sessions with tmux and SSH over VPN
- Write and edit scripts seamlessly with VSCode
- Leverage GitHub Copilot for fast prototyping and pipeline development
This post outlines my setup and some practical use cases, aimed at researchers and biologists looking to increase productivity and reproducibility.
🖥️ 1. Base Environment: WSL2 + Oh My Bash
Why WSL2?
The Windows Subsystem for Linux (WSL2) is a game-changer for bioinformatics on Windows. It provides a full Linux kernel, allowing me to run native Linux applications and tools directly on my Windows machine (without the overhead of a VM). This setup eliminates the need for dual-booting or using virtual machines, making it easy to switch between GUI applications and command-line tools. With WSL2, you can even run Linux GUI applications alongside Windows apps, providing a seamless experience.
It’s ideal for combining GUI apps (e.g., VSCode, Excel) with command-line tools (e.g., snakemake, blast, samtools). You can install WSL2 by following the official guide or even better through the Microsoft Store. You can choose Ubuntu or any other preferred distribution. You can also access your windows files from WSL2 at /mnt/c/ and see you WSL2 files in Windows in the windows file explorer.

Oh My Bash Customization
I use Oh My Bash for a cleaner prompt and quick aliases. It provides nice themes and useful aliases that speed up my workflow. I also customize my .bashrc to include aliases for common tasks, like clearing the screen, updating or checking Git status.
Oh My Bash allows you to customize your terminal prompt with themes and plugins, making it visually appealing and functional. It supports various themes, including Powerline-style prompts, which are great for displaying Git status and other information. You can also activate plugins like tmux-autoattach, which automatically attaches to existing tmux sessions for remote SSH connections.
Once Oh My Bash is installed , I also tweak my .bashrc to include useful aliases and functions. This makes my terminal experience more efficient and tailored to my workflow.
Example .bashrc snippet:
# Bioinfo aliases
alias upgrade="sudo apt update && sudo apt upgrade -y && sudo apt autoremove && sudo apt autoclean"
alias refresh="source ~/.bashrc"
alias q='exit'
alias c='clear'
alias la='ls -lah'
alias gs='git status'
alias act='conda activate'
alias srunbio='srun --mem=32G --time=4:00:00 --pty bash'
alias ..='cd ..'
alias ...='cd ../..'
alias ....='cd ../../..'
alias ..l='cd .. && ll'
alias ...l='cd ../.. && ll'
alias ....l='cd ../../.. && ll'
alias l='ls -l --color=auto'
# USAGE: mdpdf document.md document.md.pdf to convert from markdown to pdf
alias mdpdf="pandoc -s -V geometry:margin=1in -V documentclass:article -V fontsize=12pt"
# Prompt customization
OSH_THEME="agnoster"
You can also add function wrappers for reproducible workflows.
# as suggested by Mendel Cooper in "Advanced Bash Scripting Guide"
extract () {
if [ -f $1 ] ; then
case $1 in
*.tar.bz2) tar xvjf $1 ;;
*.tar.gz) tar xvzf $1 ;;
*.tar.xz) tar Jxvf $1 ;;
*.bz2) pbzip2 -d -p32 $1 ;;
*.rar) unrar x $1 ;;
*.gz) unpigz -p 32 $1 ;;
*.tar) tar xvf $1 ;;
*.tbz2) tar xvjf $1 ;;
*.tgz) tar xvzf $1 ;;
*.zip) unzip $1 ;;
*.Z) uncompress $1 ;;
*.7z) 7z x $1 ;;
*) echo "don't know how to extract '$1'..." ;;
esac
else
echo "'$1' is not a valid file!"
fi
}
# make a directory and cd into it
function mcd { mkdir -p "$1" && cd "$1";}
# Function to pretty-print CSV files in terminal
function pretty_csv {
column -t -s, "$@" | less -F -S -X -K
}
# Function to pretty-print TSV files in terminal
function pretty_tsv() {
column -t -s $'\t' "$@" | less -F -S -X -K
}
If you want more inspiration and possibilities, check out this link
🧱 2. Terminal Multiplexing with tmux
tmux is essential for working over SSH, especially with flaky VPNs (or wifi, or network connectivity issues ). It allows persistent, detachable sessions. With tmux, I can start a session, run long jobs, and detach from it (I can even shutdown my laptop). If my VPN drops or I lose connection, I can simply reattach to the session without losing progress. You don’t need to worry about losing your work if you get disconnected. Another advantage is that you can run multiple terminal windows in one SSH session, which is great for monitoring logs or running multiple commands simultaneously.
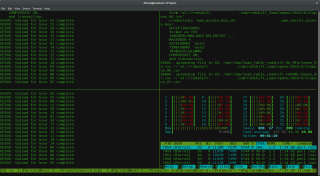
My .tmux.conf contains:
# Remap prefix to Ctrl-a
unbind C-b
set -g prefix C-a
bind C-a send-prefix
# Split panes
bind | split-window -h
bind - split-window -v
Usage example:
ssh my-hpc
tmux new -s unicycler
# Run long assembly job
If the VPN drops, I just reconnect and run:
tmux attach -t unicycler
🔐 3. SSH Access via VPN
I access clusters via VPN with openconnect, and use SSH config for shorthand:
# ~/.ssh/config
Host hpc
HostName hpc.myuniversity.edu
User steve
IdentityFile ~/.ssh/id_rsa
Then I simply use:
ssh hpc
Pro tip: use autossh for auto-reconnect.
🧠 4. VSCode for Local and Remote Editing
VSCode is my go-to editor because of:
- Git integration
- Remote extensions (
Remote - WSL,Remote - SSH), see this Youtube video to get started - Integrated terminal
- Snippets and extensions for bioinformatics (e.g., Snakemake, Biopython)
- Code completion, syntax highlighting and debugging
- Multi-language support I use the Remote - WSL extension to edit files directly in my WSL2 environment. This allows me to work with Linux tools while using a powerful GUI editor. For remote clusters, I use the Remote - SSH extension to connect and edit files directly on the server.
I often run:
code .
in WSL for native editing. For SSH:
code --remote ssh-remote+hpc /data/myproject
Sample .vscode/settings.json:
{
"python.pythonPath": "/home/steve/miniconda3/envs/bactools/bin/python",
"terminal.integrated.defaultProfile.linux": "bash",
"files.exclude": {
"**/.snakemake": true,
"**/.ipynb_checkpoints": true
}
}
My VSCode setup:
🤖 5. GitHub Copilot for Code Assistance
I use GitHub Copilot within VSCode to speed up:
- Bash scripts
- Snakemake rules
- Python functions
- R scripts
- port from other languages (for example, converting a Perl script to Python)
Copilot suggests code snippets based on context, which is especially useful for repetitive tasks or boilerplate code. It can help you quickly prototype functions, write tests, or even generate documentation.
Even the free tier of Copilot is quite powerful (and in a lot of cases enough for non-developers), providing context-aware suggestions that can save time on routine coding tasks. It learns from your coding style and adapts to your specific needs, making it a valuable tool for bioinformatics workflows.
Real example:
I start typing:
def parse_gff(file):
Copilot suggests a full parser using Biopython. I tweak it for my genome annotation workflow.
While Copilot isn’t perfect (especially on biological assumptions), it cuts down repetitive coding time significantly. You can also use Copilot to generate documentation for your scripts or functions, which is great for reproducibility and sharing your work with others. It can help you write clear and concise comments, docstrings, and even README files for your projects.
🧪 6. Real-World Workflow
Example: hybrid genome assembly using Unicycler.
# On local WSL2
code ~/projects/xanthomonas-unicycler
# Edit script with Copilot help
# SSH into cluster
ssh hpc
tmux new -s unicycler
# Launch job
unicycler -1 sample_R1.fastq -2 sample_R2.fastq -l sample_ONT.fastq -o unicycler_out
# Monitor logs in tmux pane (htop, tail -f, btop)
I version my pipeline with git, and VSCode auto-syncs changes when working over SSH.
🧭 Conclusion
This terminal setup has made my work more efficient and reproducible. Key takeaways:
- Use WSL2 to unify Linux tooling on Windows
- Leverage tmux and SSH to stay connected
- Let VSCode + Copilot speed up coding without sacrificing control
- Version control with Git for reproducibility
- Customize your environment with tools like Oh My Bash for a better experience
- Always document your workflows for future reference (let Copilot help with that too!)
Happy coding! 🧬🖥️
If you have any questions or suggestions, feel free to reach out. I’m always looking to improve my workflow and would love to hear about yours.
📎 Resources & Links
Enjoy Reading This Article?
Here are some more articles you might like to read next: